
|

|
No. 82 June 2012 |
| |
Basic Science
New Discovery on Transcript-centric Mutations
As we all start to appreciate, genetic mutations in human genomes may lead to diseases. Therefore, it is critical to understand how genetic variations arise and what these disease-causing mutations are. Dr. Cui Peng et al from the research team headed by Prof. Yu Jun from the Beijing Institute of Genomics (BIG), CAS provided novel insights into the very basic set of laws of genetic mutations in the human genome. They investigated the transcript-centric genetic variations in human genomes through analyzing data from both high-throughput RNA-sequencing in ten different tissues and single nucleotide polymorphisms in human populations. In their study, gene mutation rates were significantly and positively correlated with their expression level. This suggests that mutations may not occur randomly, but rather tend to accumulate more in genes that are expressed more frequently or at higher levels. Furthermore, they examined the frequency of 12 nucleotide mutations and found that CˇúT, AˇúG, CˇúG, and GˇúT mutations are the four major types of transcript-centric mutations in human genomes. Interestingly, they also identified a negative gradient of the sequence variations aligning from the 5' to 3' end of the transcription units (TUs), and the intensity of this gradient was correlated to gene expression levels. This phenomenon may be associated with the molecular process of transcription-coupled DNA repair (TCR) (the 5' end of TUs was more frequently repaired than the 3' end). Lastly, they presented the periodicity of genetic variations in human genome, which is supposed to be the consequence of both nucleosome phasing and TCR. Their work recently published in Genomics, Proteomics & Bioinformatics (GPB).
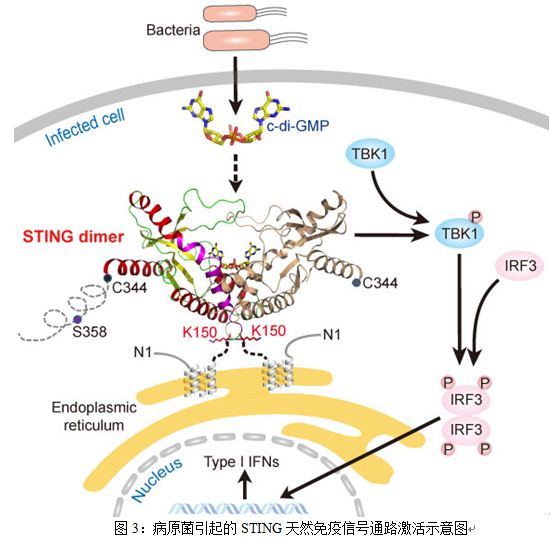
Major Achievement in STING Adaptor Protein
A stimulator of interferon genes (STING), also known as MITA, ERIS, MPYS and TMEM173, is an essential signaling molecule for DNA and cyclic di-GMP (c-di-GMP)-mediated type I interferon (IFN) production via TANK-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3) pathway. Despite the essential role of STING in DNA-mediated IFN-I induction, the mechanisms of its action are less clear and controversial in some cases. In a recent study published in Immunity, Prof. Liu Zhijie and his colleagues at the Institute of Biophysics, CAS described the crystal structures of the STING CTD alone and in a complex with c-di-GMP refined to 2.45 ? and 2.15 ? resolution, respectively. The structural, functional, and mutagenesis data unravel several hitherto unknown aspects of STING and shed light on its mechanism of activation. STING forms homotypic interactions that are essential for stimulation of IFN production. The structures of STING CTD provide detailed molecular insights into the nature of this homotypic architecture. Notably, the structure of STING CTD is very different from any known structure of adaptors or proteins functioning in innate immune responses, therefore STING may represent a novel class of sensors involved in detection of bacterial intrusion. The protein expression, analytical, and structural evidences revealed that the aa 153¨C173 region was not a transmembrane region as predicted previously but is a hydrophobic dimer interface. The structure of the binary complex maps the exact location of the c-di-GMP binding site on STING and unveils a unique mode of binding of c-di-GMP to proteins. Structure-guided mutagenesis studies showed that STING exists and functions as a dimer. In a finding contrary to previous reports, the authors show that ubiquitination of K150 may not be a prerequisite for dimerization. The c-di-GMP binding pocket is formed via dimerization of STING. Interestingly, binding of c-di-GMP enhances the association of TBK1 with STING. Although the results help with further understanding of the nature of the homotypic interactions of STING essential for stimulation of IFN production, issues on how the signal of intrusion sensed by STING is relayed, leading to recruitment of downstream effector molecules, remain to be investigated.
Significant Advance in Research on Feather Evolution
It has been assumed that the dinosaurs would have lost feathers when they became really gigantic. A research team led by Prof. Xu Xing at the Institute of Vertebrate Paleontology and Paleoanthropology, CAS recently reported a new gigantic feathered dinosaur named ˇ®Yutyrannus hualiˇŻ from the Early Cretaceous of Liaoning, China. This new finding greatly expands the known body size of feathered dinosaurs and demonstrates that the evolution of integumentary structures might be more complicated than previously realized, especially when considered in relation to gigantism. The team inferred that the presence of extensive feathers in the gigantic Yutyrannus huali was probably linked to the relatively cold environment that prevailed approximately 125 million years ago, when such dinosaur lived. A previous study based on oxygen isotopic analysis of dinosaur teeth indicated that temperatures at this time were significantly lower than later periods of the Cretaceous, which was generally a warm time in the EarthˇŻs history. The plumage of Yutyrannus huali thus suggests that the development of feathers is related not only to body size, but also to temperature. This new discovery has attracted considerable attention from the international scientific community. Dr. Mark Norell of the American Museum of Natural History commented that ˇ°the discoveries of feathered dinosaur fossils from China have completely changed our conventional knowledge of dinosaursˇ±. This study was published in Nature on April 5, 2012.
copyright © 1998-2015
CAS Newsletter Editorial Board: 52, Sanlihe Road, Beijing 100864,
CHINA
Email: slmi@cashq.ac.cn