
|

|
No. 81 April 2012 |
| |
Bioscience
Interaction between KSHV LANA and RBP-Jk: Important for Viral Latency
Kaposi¡¯s sarcoma-associated herpesvirus (KSHV) is a recently discovered human tumor virus, which belongs to the gamma-2 herpesvirus subfamily. Following primary infection, the default pathway of KSHV infection is latency. However, the mechanism of which KSHV establishes latency is not fully understood. Thus, to further understand the maintenance mechanism of viral latency, Jin Yi and her colleagues, supervised by Prof. Lan Ke at Institut Pasteur of Shanghai, CAS, identified a motif in LANA C-terminal, which is responsible for LANA interaction with RBP-J¦Ê. By reconstitution of the virus and analysis of the function of motif deficient LANA in the context of the full-length KSHV genome, they found that the interaction between LANA and RBP-J¦Ê was important for repressing viral lytic replication and maintaining KSHV latency. Their findings provide a valuable evidence for understanding the regulation of viral genes by a host transcription factor and its potential role in maintaining KSHV latency. It could be a potential target for disruption of viral latency, which could be a strategy for oncolytic viral therapy. This work was published online Journal of Virology on February 29, 2012.
ParaAT: A Parallel Tool for Constructing Multiple Protein-coding DNA Alignments
Prof. Zhang Zhang and his team at the Beijing Institute of Genomics (BIG), CAS, have developed a parallel tool for constructing multiple protein-coding DNA alignments called ParaAT (Parallel Alignment & back-Translation), which successfully overcomes the disadvantages of traditional tools for protein-coding nucleotide alignments. ParaAT achieves parallel construction of protein-coding DNA alignments for a large number of homologs. It is not only suitable for large-scale data analysis in generating multiple protein-coding nucleotide alignments, but also capable of reducing the running time and exhibiting nearly linear speedup. ParaAT is provided as open-source software and can be executed on Linux/Unix, Macintosh, and Windows platforms. It supports different output formats, which allows further related bioinformatic analysis, such as detection of natural selection by KaKs_Calculator. This study was published in a recent issue of Biochemical and Biophysical Research Communications.
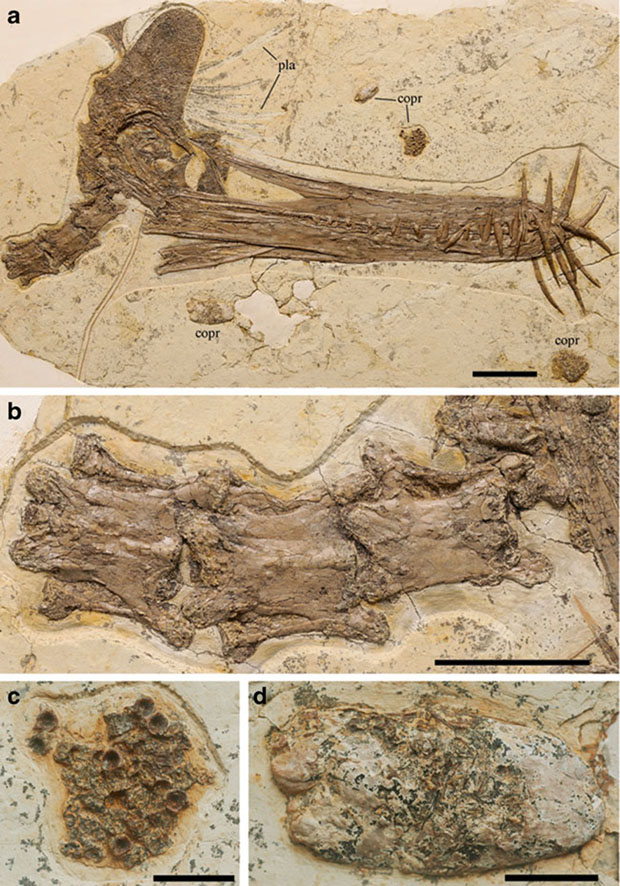
Guidraco venator : A New Cretaceous Pterosaur Found
A new special pterosaur fossil, Guidraco venator, was discovered in Liaoning, China. Its head shape is reminiscent of a malevolent ghost and its huge teeth of the anterior tips indicate the professional hunting ability, for which it was named. G. venator and Ludodactylus from Brazil have many similarities. This discovery proves that the pterosaur fauna from the Jiufotang Formation (Lower Cretaceous) of western Liaoning is very similar to that from the Santana Formation (including Crato and Romualdo formations or members) of the Araripe basin in northeastern Brazil, and that they have a close evolutionary relationship. G. venator was found in the lacustrine shale of the late Early Cretaceous (120 Ma) in Sihedang locality of Lingyuan City. It is a new vertebrate fossil locality of Jiufotang Formation in western Liaoning. This specimen preserved a complete and articulated skull and the anterior portion of a neck. G. venator has a 38 cm skull with following characters: nasoantorbital fenestra length about one fourth total skull length; high, casque-shaped frontal crest with subperpendicular anterior margin, and rounded dorsal margin; rostral teeth robust, large and inclined anteriorly, with crowns surpassing the margins of the skull during occlusion. Prof. Wang Xiaolin and his doctoral students from the Institute of Vertebrate Paleontology and Paleoanthropology, CAS, working with a Brazilian paleontologist Alexander Kellner, published their result titled New toothed flying reptile from Asia: close similarities between early Cretaceous pterosaur faunas from China and Brazilin Naturwissenschaften( Apr. 2012, Vol. 99, No. 4, pages 249-257).

copyright © 1998-2015
CAS Newsletter Editorial Board: 52, Sanlihe Road, Beijing 100864,
CHINA
Email: slmi@cashq.ac.cn