
|

|
No. 94 April 2014 |
| |
Bioscience
A New Gene for Human Obesity Discovered
A research group headed by Prof. Li Wei at the Institute of Genetics and Developmental Biology, CAS has been studying the mechanism of vesicle trafficking disorders and diseases. It is understood that a locus on chromosome 6q23-25, D6S1009, was linked to obesity or body mass index (BMI). The group, by observing SLC35D3 deficient mice at the side of the locus, discovered the characteristics of progressive obesity and the metabolic syndrome in the mice of two months old (equals to human adults). Further, they discovered the incidence of obesity caused by affecting of the membrane trafficking of dopamine receptor D1 (D1R) and impairing of dopamine signaling in striatal neurons of basal ganglia due to the defected encoding protein of this gene. The symptom of obesity and metabolic syndrome is recovered to normal standard when the obese mice is treated by D1R agonist. The research result demonstrates that SLC35D3 is the disease-causative genes for human obesity and metabolic syndrome. Relevant result was published in magazine PLoS Genetics on February 13, 2014.
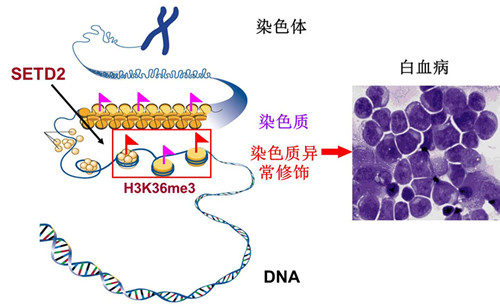
New Discovery Reveals a Tumor Suppressor Gene in Acute Leukemia
Recently, the lab of Prof. Wang Qianfei from the Laboratory of Genome Variations and Precision Bio-Medicine in Beijing Institute of Genomics, CAS revealed functional cooperative mutations of SETD2 in acute leukemia, in collaboration with Profs. Cheng Tao and Zhu Xiaofan from the Institute of Hematology and Blood Diseases Hospital and Center for Stem Cell Medicine, Chinese Academy of Medical Sciences and Peking Union Medical College, and Prof. Gang Huang from Cincinnati Children’s Hospital Medical Center in USA. This work has been online published by Nature Genetics, which is titled as “Identification of functional cooperative mutations of SETD2 in human acute leukemia”.They found that abnormal chromatin modifications can promote acute leukemia, and SETD2 is a new tumor suppressor gene in acute leukemia.
Through whole genome sequencing, RNA sequencing and related bioinformatics analyses, Dr. Wang and colleagues identified an oncogenic MLL-NRIP3 fusion gene and mutations in SETD2, a key histone modifying enzyme gene that is responsible for catalyzing trimethylation of H3K36 (H3K36me3). Loss-of-function point mutations in SETD2 were recurrent (6.2%) in 241 patients with acute leukemia and were associated with multiple major chromosomal aberrations. Disruption of the SETD2 function resulted in global loss of H3K36me3 in leukemia cells. In the presence of a major chromosomal translocation, SETD2 disruption greatly accelerated both initiation and progression of leukemia, via enhancing the self-renewal potential of leukemia stem cells (LSC) and activating multiple signaling pathways. Moreover, specific inhibition of the mTOR pathway significantly decreased the self-renewal potential of LSC.Therefore, this study provides compelling evidence for SETD2 as a new tumor suppressor. Disruption of the SETD2-H3K36me3 pathway is a distinct epigenetic mechanism for leukemia development.Whole genome sequencing in conjunction with the series of experimentation demonstrated that chromatin modification abnormality caused by SETD2 mutations is a promising molecular therapeutic target. The distinct epigenetic mechanism offers a new opportunity for clinical prediction, diagnostics and therapeutics of acute leukemia.
Breakthrough in Biodiversity Monitoring
Douglas Yu’s group at KIZ reports that biodiversity can be cataloged and monitored by bulk DNA sequencing of insect specimens. They recently have published this novel biodiversity monitoring technology — called metabarcoding — in Ecology Letters. This technology is faster and more reliable than the traditional methods used by many taxonomy experts, which require expert identification of species. The biggest advantage of metabarcoding is that it can be used by people who are not taxonomic experts, allowing them to detect environmental change and to detect the status of endangered or pest species. This technology is already being applied in numerous locations. For instance, metabarcoding is being used to search for tree kangaroos in Papua New Guinea and to detect moth community change with climate change. Metabarcoding can also be used to monitor the progress of ecological restoration experiments, such as the restoration of rubber plantations in China, heathland in the UK, and oil palm plantations in Sumatra.
copyright © 1998-2015
CAS Newsletter Editorial Board: 52, Sanlihe Road, Beijing 100864,
CHINA
Email: slmi@cashq.ac.cn