

A study published in Science Advances on August 17 revealed a widespread length dependent splicing regulation in cancer. By analyzing various transcriptome data from The Cancer Genome Atlas (TCGA), this study discovered a common feature of cancer-specific splicing dysregulation and provided important clinical implications for cancer diagnosis and therapies.
Over 95 percent of human genes undergo alternative splicing (AS), generating multiple mRNA isoforms with distinct functions from a single gene. The widespread splicing dysregulation is one of the molecular hallmarks of cancer, and targeting the mis-spliced genes may be a powerful therapeutic strategy against it. Therefore, systematic study of AS in cancer is critical for precision cancer medicine. While several studies provided useful information by analyzing various transcriptome data, a general trend in cancer-associated splicing dysregulation has not yet been determined and the underlying mechanisms remain unclear.
In this study, a comprehensive analysis of AS changes using transcriptome data from thousands of patients with 18 types of cancers revealed an unexpected length dependency in cancer-associated exons. Compared to typical exons, the short exons are more likely to be mis-spliced and preferentially excluded in almost all cancers. These cancer-associated short exons (CASEs) are more conserved, and more likely to encode in-frame peptides and functional enrichment in GTPase regulators and cell adhesion. The researchers further developed machine learning algorithms with CASEs as diagnostic markers and defined a CASE-based risk factor to accurately predict the prognosis of cancer patients. Finally, they determined the possible mechanisms for this length-dependent regulation, which is distinguished by an elevated transcription rate and alteration of certain RNA binding proteins in cancers.
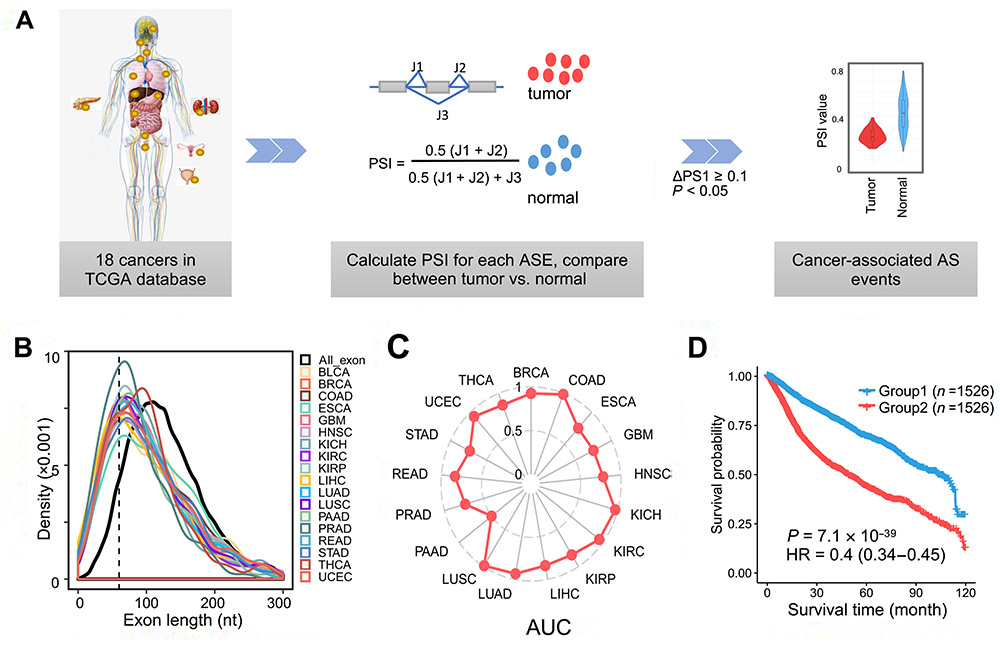
(A) Schematic diagram to identify cancer-associated ASEs. (B) The length distribution of all human exon and cancer-associated exons in each cancer type. (C) The ROC curve of the random forest model in each cancer type. (D) The Kaplan-Meier curve of cancer patients grouped by the CASE-based risk factor. [IMAGE: DR. WANG ZEFENG’S GROUP]
Collectively, this study reveals a general rule for splicing dysregulation in cancers, provides a simple and practical route for cancer stratification, and uncovers the potential underlying mechanisms. Since the splicing dysregulation was recently found to be associated with cancer immunotherapy and drug assistance, this study provides valuable information for cancer prediction with clinical implications.
The research, entitled “A widespread length-dependent splicing dysregulation in cancer”, was published online in the journal of Science Advances on August 17. Dr. Wang Zefeng from the Shanghai Institute of Nutrition and Health (SINH) of the Chinese Academy of Sciences (CAS) and Dr. Wang Yang from Dalian Medical University are the co-correspondent authors of this work. Dr. Zhang Sirui from CAS’s SINH is its first author.
For more information, please contact:
Wang Jin
E-mail: wangjin01@sinh.ac.cn
Shanghai Institute of Nutrition and Health (SINH),
Chinese Academy of Sciences
Source: Shanghai Institute of Nutrition and Health (SINH),
Chinese Academy of Sciences