

On September 23, 2020, GMT, the Xu Huaqiang group from the Shanghai Institute of Materia Medica (SIMM), Chinese Academy of Sciences, together with the Karsten Melcher group and the Peter Jones group from the Van Andel Institute (VAI) of the United States published important research on de novo DNA methyltransferase (DNMT) entitled “Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B” in Nature.
The teams used advanced Cryo-EM technology to determine the high-resolution structure of de novo DNMT3A2/DNMT3B3 with nucleosome substrate, elucidated the binding mode of DNMT3A2/DNMT3B3 with a nucleosome core particle, and proposed a genome-wide DNA methylation model.
DNA methylation can change chromatin structure, DNA stability, and DNA-protein interactions, which can control gene expression. DNA methylation can be inherited by the offspring DNA along with the DNA replication process which makes it an important epigenetic mechanism. The DNA methylation in a chromatin context is far more complex than in solution, particularly because the DNA wrapped by a nucleosome is refractory to cytosine modification by either prokaryotic or mammalian enzymes. Also, most of the de novo DNMT3s bound to nucleosomes are not actively engaged in catalysis. CpG methylation by the de novo DNMT3A and DNMT3B is essential for mammalian development and differentiation and is frequently dysregulated in cancer.
By analyzing the expression patterns of different subtypes of DNMTs in thousands of normal tissues (GTEx database) and cancerous tissues (TCGA database), this study focused on determining the interaction of DNMT3A2 and DNMT3B3 with a nucleosome core particle (NCP), because these are the two main subtypes expressed in human cancers. The crystal structures of isolated DNMT3A catalytic domain and DNMT3L catalytic like domain with or without free DNA have been analyzed. However, due to its limitations, the interaction mechanism between DNMT and its natural substrate nucleosome has not been illustrated.
To reveal the interaction between DNMT3A2/3B3 and nucleosomes, and to understand the DNA methylation on chromatin, the joint group successfully determined in near-atomic resolution the cryo-EM structure of nucleosome-bound DNMT3A and DNMT3B complexes.
The structure shows that a heterotetrameric 3B3-3A2-3A2-3B3 complex is very similar to that formed by the isolated catalytic domains and catalytic-like domains of DNMT3A and DNMT3L. This complex interacts asymmetrically with the NCP, with one of the two DNMT3B3 catalytic-like domains anchored to the acidic patch region of the nucleosome core. Central to this interaction are the DNMT3B3 arginine finger residues R740 and R743. This acidic patch is a key structural element that also has crucial interactions with many other NCP-binding proteins. The DNMT3A2 catalytic domains do not contact the nucleosome core but rather follow the path of the linker DNA, interact with the linker DNA and methylate the linker DNA. Despite the conserved catalytic domains in the DNMT3 family, structural alignment of DNA-bound DNMT3A2 and nucleosome core-bound DNMT3B3 revealed a switch function for the TRD. The TRD found in all catalytically active DNMT3A and 3B subtypes makes key contributions to DNA binding, yet sterically blocks the binding of the catalytic domains to the nucleosome core.
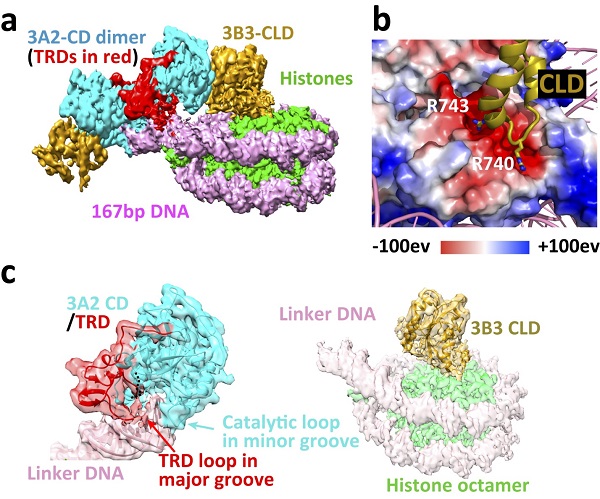
a. Cryo-EM density map of the human DNMT3A2-DNMT3B3-NCP complex
b. Acidic patch interaction
c. Binding of the DNMT3A2 catalytic domain (3A2 CD) to linker DNA and binding of the DNMT3B3 catalytic-like domain (3B3 CLD) to the histone core [IMAGE: XU TINGHAI]
To test the importance of the acidic patch interaction for nucleosome recruitment, mutation analysis at the center positions of 740 and 743 of 3B3, and amino acids 745 and 749, which are away from the acid patch region of nucleosomes as controls, was performed.
In vitro interaction experiments (ALPHA Screen) showed that the opposite charge mutations near the acidic patch (K745E or R749E) had little influence, while R740E and R743E at the acidic patch strongly reduced the ability of the DNMT3A2/DNMT3B3 heterotetramer to bind the NCP. As expected, the four same-charge mutations showed no significant changes in binding ability.
In cell chromatin association assay also confirmed that acidic patch interaction residues 740 and 743 arginine mutations lead to significantly reduced binding to chromatin. To test whether the acidic patch interaction is important for reestablishing DNA methylation patterns in vivo, DNA methylation (Infinium MethylationEPIC BeadChip) was analyzed.
The analysis revealed that the ability of mutant DNMT3B3 to restore DNA methylation is related to chromatin binding capacity. The K745 and R749 mutant methylation, as controls, showed similar methylation recovery patterns as wild-type DNMT3B3. The limited micrococcal nuclease (MNase) digestion assay confirmed the strong increase in protection from nuclease digestion of about 10 bp on either side of the NCP in the presence of the DNMT complex. These data strongly indicate the importance of the interaction of the catalytic-like domain of DNMT3B3 with the acidic patch for nucleosome recruitment and DNA methylation, and that the binding to DNA does not require a CpG at the active site of the enzyme.
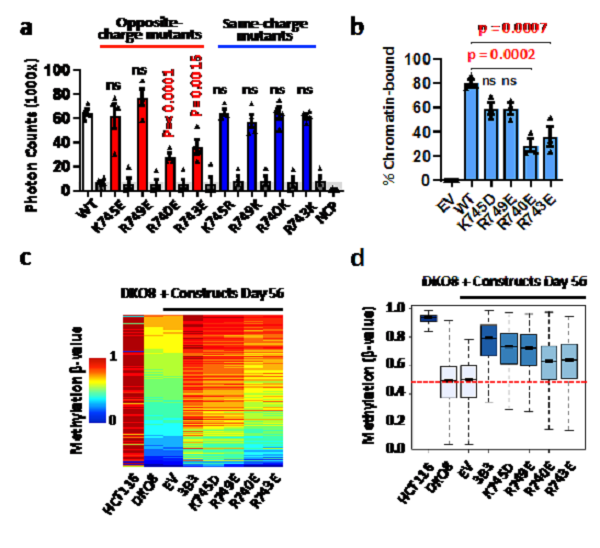
a. AlphaScreen interaction assay between His-tagged DNMT3A2–DNMT3B3 complexes and biotinylated NCP
b. Graphical representation from three independent biological replicates of DNMT3 chromatin association assays
c. Methylation heatmap of CpG sites in wild-type (HCT116) cells and DKO8 cells expressing DNMT3B3 WT or CLD mutants
d. Box plot of 109,998 probes, showing the distribution of DNA methylation levels for each cell line [IMAGE: XU TINGHAI]
By separating core nucleosome targeting and CpG methylation through catalytically inactive and active DNMTs, the DNMT3 complex can be recruited to core nucleosomes with its poorly accessible DNA while targeting CpG methylation to accessible linker DNA. This suggests that remodeling of the nucleosome core is required for spreading DNA methylation to nucleosomal DNA in vivo, e.g., through DNA replication, transcription, or other nucleosome remodeling events.
This work was initiated by Xu Tinghai, who is the first author of this article. Xu Tinghai got doctorate degree in 2017 from SIMM, and currently is a postdoctoral fellow jointly supervised by Dr. Peter Jones and Dr. Karsten Melcher at VAI.
Dr. Liu Minmin, Dr. X. Edward Zhou, Dr. Zhao Gongpu, and Professor Liang Gangning from the University of Southern California involved also in this work. Dr. Xu Huangqiang from SIMM, and Dr. Peter Jones, Dr. Karsten Melcher from VAI are the co-corresponding authors.
For more information, please contact:
Prof. Xu Huangqiang
Email: eric.xu@simm.ac.cn
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
Source: Shanghai Institute of Materia Medica,
Chinese Academy of Sciences