

Research teams led by Dr. Yang Li at the CAS-MPG Partner Institute for Computational Biology, Chinese Academy of Sciences, Dr. Chen Jia at the School of Life Science and Technology, ShanghaiTech University and Dr. Shen Bin at Nanjing Medical University demonstrate the underlying mechanism of mutagenesis during repair of the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated 9 (Cas9)-generated DNA breaks, providing new strategies to increase the fidelity and accuracy of CRISPR/Cas9-mediated genome editing.
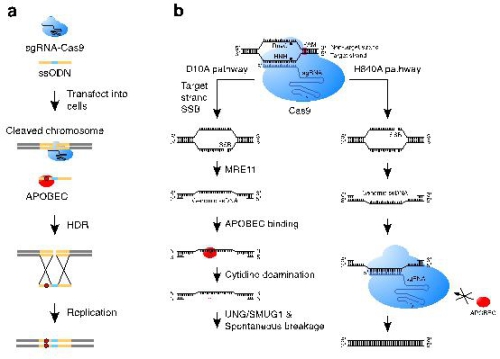
CRISPR/Cas9 system, which provides so far the most convenient and efficient genome editing, has been broadly applied in life science research and shows great potential in clinics to treat human diseases. However, due to the risk of permanent and inherited off-target/unintended editing in genomic DNA, scientists strive to improve CRISPR technology, aiming to increase its fidelity and accuracy. The Apolipoprotein B mRNA editing enzyme, of the catalytic polypeptide-like/activation-induced cytidine deaminase (APOBEC/AID) family of proteins, prefers single-stranded nucleic acids for cytidine to uracil deamination. Single-stranded nucleic acids, such as single-stranded oligo deoxy nucleotides (ssODNs) and genomic single-stranded DNA (ssDNA), are commonly involved in the DNA repair system for the breaks generated by CRISPR/Cas9. Thus, it is intriguing to examine whether APOBECs can target these single-stranded nucleic acids to affect the repair outcome of CRISPR/Cas9-generated DNA breaks.
In this current study, researchers showed that APOBEC3 can trigger cytidine deamination of ssODN, which ultimately results in base substitution mutations in genomic DNA through homology-directed repair of Cas9-generated double-strand breaks. In addition, the APOBEC3-catalyzed deamination in genomic single-stranded DNA formed during the repair of Cas9 nickase-generated single-strand breaks in human cells can be further processed to yield mutations mainly involving insertions or deletions (indels). Both APOBEC3-mediated deamination and DNA-repair proteins play important roles in the generation of these indels. Based on these findings, researchers suggest using dsODN, instead of ssODN, as homologous donors for high-fidelity gene correction in postnatal tissues or organs, and temporarily suppressing endogenous APOBEC3s to repress these unwanted mutations in genomic DNA.
The paper entitled “APOBEC3 induces mutations during repair of CRISPR–Cas9-generated DNA breaks” was published online in Nature Structural & Molecular Biology on Dec 11, 2017.
For more information, please contact:
Prof & Dr. Yang Li
Computational Transcriptomics and Bioinformatics
CAS-MPG Partner Institute for Computational Biology
Shanghai Institutes for Biological Sciences
Chinese Academy of Sciences
E-mail: liyang@picb.ac.cn
Source: Shanghai Institutes for Biological Sciences, CAS