

A team of scientists led by Professor Liu Zhijie at the Institute of Biophysics, Chinese Academy of Sciences has determined the 3-dimensional molecular structure of the agonist-bound human cannabinoid receptor CB1. The work reveals the structural features of agonist-bound CB1 and the activation mechanism of the receptor. The results, described in a paper entitled Crystal structures of agonist-bound human cannabinoid receptor CB1, were published online on July 6, 2017 in journal Nature.
G protein-coupled receptors (GPCRs) are the largest and most diverse protein family in the cell membrane of the human body and play very important roles in signal transduction. About 40% of drugs on the market target GPCRs. The agonists will activate the receptor, while the antagonists will inhibit the activation of the receptor. Both agonists and antagonists show potential for drug discovery and medication.
CB1 is one of the most highly expressed GPCRs in the human body and is the target of one of the longest used herbs, marijuana. Its ligands, including agonists and antagonists, can be used for treatment of pain, neurodegenerative disease, obesity, and liver fibrosis. Last year, the iHuman team solved the 3D structure of CB1 in complex with antagonist AM6538 which and published their results in Cell. The work revealed the structural features of the antagonist-bound CB1 and the binding modes of the antagonists, yet does not inform us as to how the agonists like THC which is the major component of marijuana bind to the receptor and how CB1 elicits its diverse physiological effects.
“Crystal structures of active state G protein-coupled receptor (GPCR) have been challenging to obtain due to their increased conformational flexibility and resulting instability,” said Dr. Hua Tian, the first author of this paper. With a great deal of effort, the team finally solved the crystal structures of CB1 bound to the two novel agonists AM11542 and AM841 designed, synthesized and analyzed by Professor Alex Makriyannis group at Northeastern University and characterized by the Professor Laura Bohn group at Scripps Florida.
“We are thrilled to see that the agonists AM11542 and AM841 could significantly stabilize the conformation of the receptor and lead to the structure determination. The two CB1-agonist complexes reveal significant conformational changes in the overall structure, relative to the antagonist-bound state, including a 53% reduction in volume of the ligand binding pocket and an increase in the surface area of the G protein binding region,” said Prof. Liu Zhijie, the lead corresponding author of this paper.
“In addition, we observed a 'twin toggle switch' of Phe2003.36/Trp3566.48 which appears to be essential in CB1 activation,” said Dr. Zhao Suwen, a collaborator at iHuman Institute of Shanghai Tech University. “The structures also provide a molecular basis for predicting the binding modes of THC, the endogenous and synthetic cannabinoids and reveal important insights into the activation mechanism of CB1.”
For more information, please contact:
Liu Zhijie
Principal Investigator (guest)
National Laboratory of Biomacromolecules
Institute of Biophysics, Chinese Academy of Sciences
iHuman Institute, Shanghai Tech University
E-mail: liuzhj@shanghaitech.edu.cn
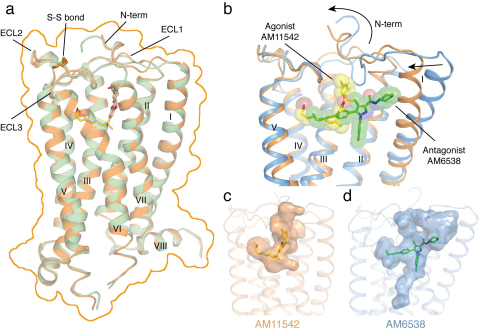
Figure: Overall structures of CB1–AM11542 and CB1–AM841 complexes and agonist-bound (orange cartoon) and antagonist-bound (blue cartoon) CB1 ligand binding pockets. Agonist AM11542 (yellow) and antagonist AM6538 (green) are shown in sticks and sphere representations. (Image by IBP)
Source: Institute of Biophysics, CAS