

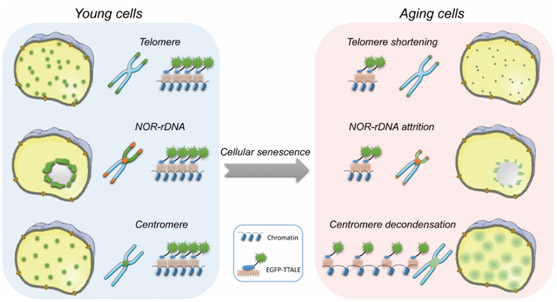
Visualization of specific genomic loci in live cells is a prerequisite for the investigation of dynamic changes in chromatin architecture during diverse biological processes, such as cellular aging. However, current precision genomic imaging methods are hampered by the lack of fluorescent probes with high specificity and signal-to-noise contrast. We find that conventional transcription activator-like effectors (TALEs) tend to form protein aggregates, thereby compromising their performance in imaging applications. Through screening, we found that fusing thioredoxin with TALEs prevented aggregate formation, unlocking the full power of TALE-based genomic imaging. Using thioredoxin-fused TALEs (TTALEs), we achieved high-quality imaging at various genomic loci and observed aging-associated (epi) genomic alterations at telomeres and centromeres in human and mouse premature aging models. Importantly, we identified attrition of ribosomal DNA repeats as a molecular marker for human aging. Our study establishes a simple and robust imaging method for precisely monitoring chromatin dynamics in vitro and in vivo.
In the human nuclear genome, ~3.2 billion base pairs of DNA are tightly packed into 23 chromosome pairs of varying size. Although we can obtain sequence information for any genomic locus with ease, we are still in the early stages of unraveling the organization of the human genome and the spatiotemporal relationships between genomic loci in three-dimensional (3D), which will consequently improve our understanding of genetic and epigenetic regulation during cell differentiation, aging, and pathophysiological processes. The importance of maintaining a proper 3D genome topology is underscored by recent discoveries that aging and several severe human diseases can be attributed to genomic disorganization. Therefore, exploring the spatial organization of human chromatin and its dynamic interactions with protein and RNA regulators emerges as an important part of understanding the cellular processes underlying aging and human diseases.
Despite recent advances, there is a lack of imaging tools suitable for accurate and long-term tracking of topological changes at specific genomic loci. A series of tools have been reported for visualization of repetitive DNA sequences or specific gene loci in fixed or live human cells, but they suffer from several limitations. (1) Fluorescent Lac or Tet operator and repressor systems have been used to visualize target genomic sequences in live cells, but the integration of large artificial sequences (~10 kb) into the genomic region of interest suffers from low efficiency and a risk of disturbing genomic structures. (2) Fluorescent in situ hybridization (FISH) has been widely used to study nuclear localization of specific sequences and genomic aberrations, but can only be performed on fixed cells after DNA denaturation. (3) Recently, the CRISPR/dCas9 system has been adapted for visualization of specific genomic loci (e.g., protein-coding mucin genes such as MUC4) and repetitive telomere sequences in live cells. However, low signal-to-noise ratios, inefficient delivery of the bulky Cas9 and single-guide RNA (sgRNA) constructs, and potential cellular stresses caused by overloading sgRNA have limited their utility. (4) Fluorescent protein-labeled transcription activator-like effectors (TALEs) have been used for live imaging of genomic repetitive sequences. TALEs are proteins secreted by Xanthomonas bacteria that infect various plant species. TALEs are DNA-binding proteins that contain tandem 33- to 35-amino acid (aa) repeats, each of which specifically recognizes and binds to a single target DNA base. The smaller size of TALEs and simple correlation between TALEs and target DNA bases makes them extremely useful for designing artificial constructs capable of recognizing genomic sequences in diverse experimental systems. Indeed, engineered TALEs have been harnessed for a variety of applications, including genome editing (when fused to the cleavage domain of FokI nuclease or to meganucleases) and design of customized transcriptional modulators and recombinases. Due to their relatively small size, fluorescently tagged TALEs have been used as small protein probes to track specific genomic DNA sequences, especially within telomeres and centromeres, in live cells. Despite these advances, a careful validation of TALE-based imaging in different cellular systems is still needed. Importantly, TALE- and Cas9/sgRNA-based imaging systems have seldom been tested in physiological and pathological contexts such as human aging.
Here we report that conventional TALEs frequently form large aggregates in human cells, thereby compromising their imaging efficiency in various cell types examined. To overcome this barrier, we developed a novel thioredoxin-fused TALE (TTALE) imaging system that can effectively eliminate aggregates and enable high-contrast visualization of the 3D dynamics of specific genomic structures under diverse physiological and pathological contexts (e.g., aging) across a wide range of cell types in vitro and in vivo.
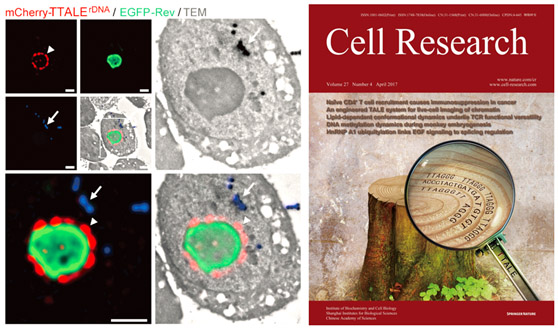
Precise visualization of telomeres and centromeres using TTALEs was finally realized. To adapt TALEs for visualizing specific genomic locus, we generated an EGFP-tagged TALE construct targeting a 19-bp telomeric DNA repeat (5′-TAACCCTAACCCTAACCCT-3′; referred to as TALEtelo). This construct was transiently introduced into four human tumor cell lines (U2OS, HeLa, HepG2, and MCF7). To examine the efficiency of TALE-based fluorescence imaging of telomeric sequences, 3D-FISH with a telomeric peptide nucleic acid (PNA) probe was used to visualize the 3D distribution of telomeres inside the nucleus. Approximately 20% of FISH-positive foci overlapped with EGFP-TALEtelo in U2OS cells. In other cell types examined, EGFP-TALEtelo appeared mostly as large bright foci (referred to hereafter as “aggregates”) distinct from telomeric foci, with only a small percentage (< 5%) of foci co-localized with the FISH signals (Figure 1A and 1C). In addition, an EGFP-tagged TALE against a 19-bp centromeric satellite DNA repeat (5′-TCCATTCCATTCCATTCCA-3′), referred to as TALEcentro, also failed to faithfully identify centromeres in the same cell types, as determined by 3D-FISH with a centromeric PNA probe (Supplementary information, Figure S1A and S1B). Similar results were also observed using previously reported EGFP-TALEs against a 15-bp telomeric repeat (5′-TAACCCTAACCCTAA-3′; Figure 1B and 1C) or a 20-bp centromeric repeat (5′-TAGACAGAAGCATTCTCAGA-3′; data not shown). We further verified that these aggregates were not localized in nucleoli (Supplementary information, Figure S1C-S1D). These data indicate that conventional TALEs are inefficient for tracking human telomeric and centromeric DNAs. The result has laid a solid foundation for understanding the nature of the human aging system based on genetics and epigenetic level. Relevant technique has been applied for patent of innovation. The progress was highly commented on by international colleagues.
The achievement was published in Cell Research on April 7, entitled Visualization of Aging-Associated Chromatin Alterations with an Engineered TALE System as its cover story, by research groups headed separately by Prof. Liu Guanghui and Xu Tao at the Institute of Biophysics, the Chinese Academy of Sciences. Professors Liu Guanghui and Xu Tao from the Institute of Biophysics and Professor Qu Jing from the Institute of Zoology are co-corresponding authors, Associate Professor Ren Ruotong, PhD student Deng Liping and Senior Engineer Xue Yanhong are first authors of the paper. This achievement was made through collaboration of researchers from different institutions, including the Institute of Biophysics, Institute of Zoology, Peking University Third Hospital, the MOH Key Laboratory of Geriatrics, Beijing Hospital, National Center of Gerontology, Department of Pediatrics, Beijing Shijitan Hospital Capital Medical University, the Institute of Aging Research, and the Hangzhou Normal University School of Medicine.
For more information, please contact:
Professor Liu Guanghui, E-mail: ghliu@ibp.ac.cn; Professor Qu Jing, E-mail: qujing@ioz.ac.cn; Professor Xu Tao, E-mail: xutao@ibp.ac.cn
Source:
1 National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China
2 State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China